According to new atlas, the algorithm helps to find a new antibiotic candidate drug, which is expected to be used against some drug-resistant bacteria . It uses a new attack mode, making it difficult for bacteria to develop drug resistance. The most important thing is that it can open up a new antibiotic library.
We humans are not the only organisms that want to kill bacteria - nature has produced a wide range of antibacterial compounds, many of which are used by bacteria themselves to gain the upper hand in the long "turf war" with other bacteria.
Most of the current antibiotics come from this "arsenal", originally grown from bacterial cultures and later synthesized into more powerful forms. The problem is that over time, bacteria become resistant to these drugs, forcing people to make new drugs. In recent decades, antibiotic resistant bacteria have become one of the most urgent health threats in the world.
Sean Brady, corresponding author of the study, said: "many antibiotics come from bacteria, but most bacteria cannot grow in the laboratory. Therefore, we may have missed most antibiotics."
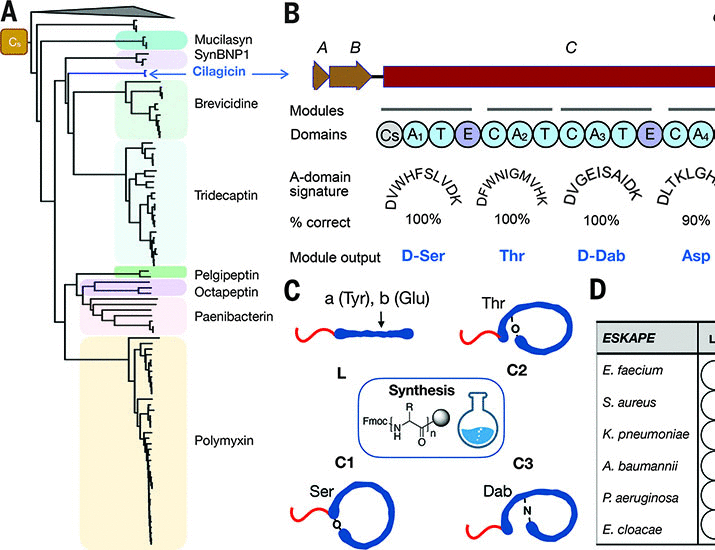
To help resolve the possibilities more quickly, Rockefeller University researchers used algorithms to investigate so-called biosynthetic gene groups. These are genomes that encode a range of proteins -- including some that may have antibacterial properties -- but for humans, they are too numerous and cumbersome to classify.
"Bacteria are complex. Just because we can sequence a gene doesn't mean we know how bacteria turn it on to produce proteins. There are thousands of undetermined gene clusters, and we only know how to activate a small part of them," Brady said But the algorithm can classify these gene clusters faster and select the most promising candidate compounds with antibacterial effect. From there, human chemists can synthesize the much shorter list of compounds and test them.
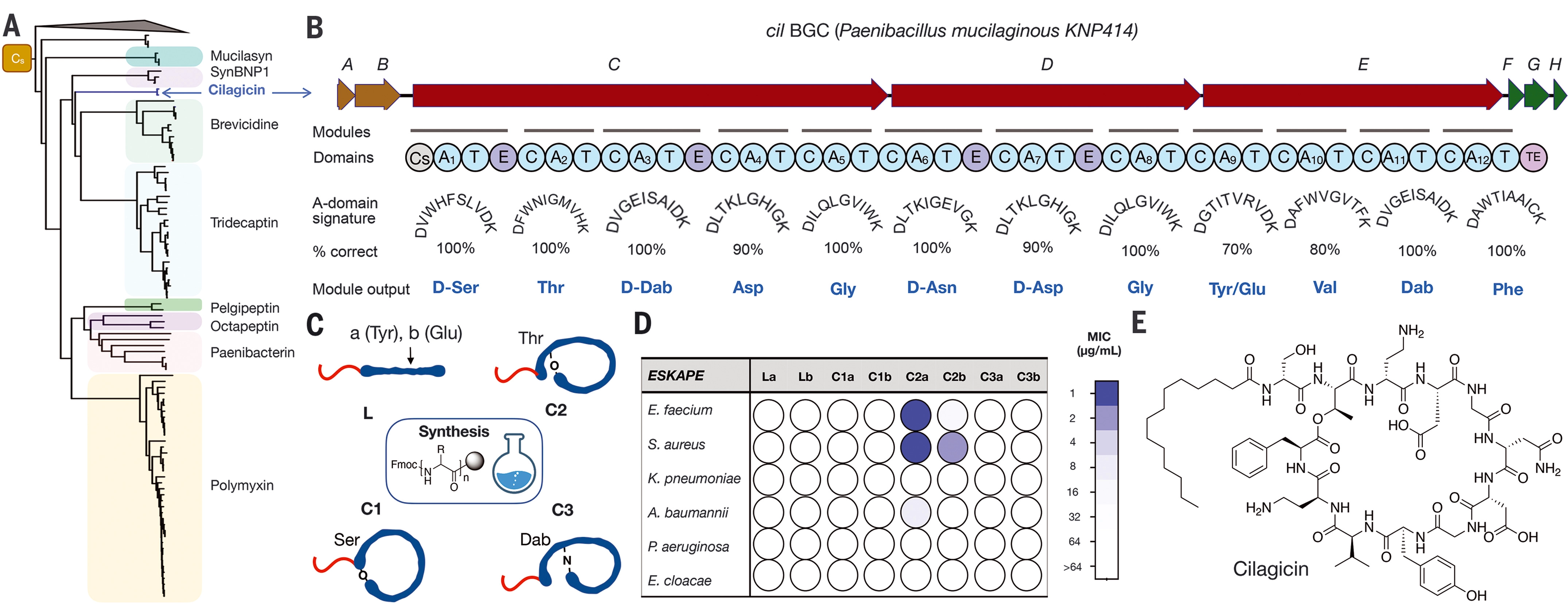
Sure enough, this process produced a particularly promising compound, which the team named cilagicin. It originated from a genome called "CIL" and was selected because it is similar to other antibiotic producing genes.
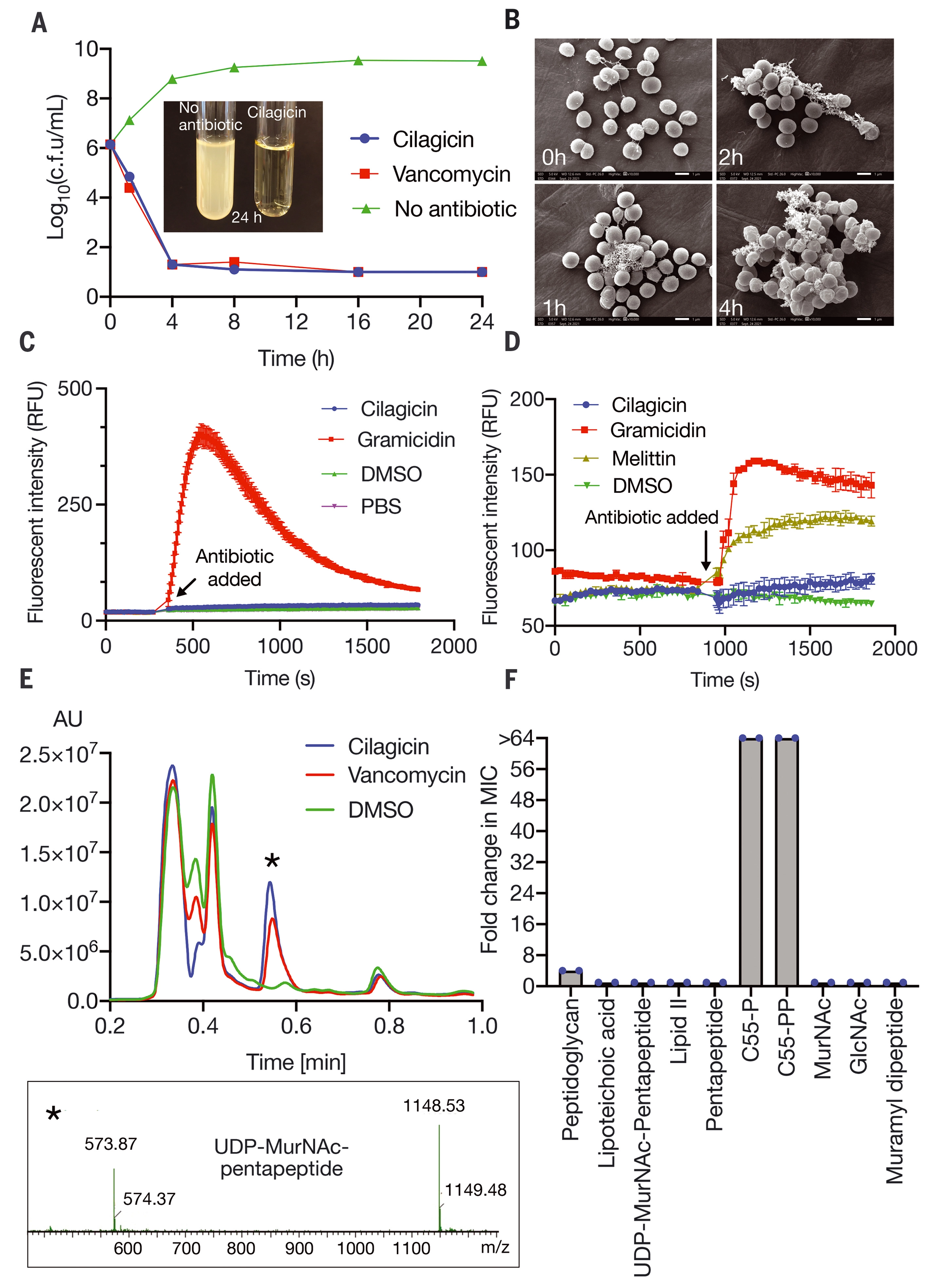
In laboratory tests, cilagicin can reliably kill bacteria, including several strains resistant to existing antibiotics. Importantly, it does not harm human cells and can treat bacterial infections in living mice. However, the most impressive thing is that it can even kill the bacteria specially designed by researchers to resist the drug.
After careful examination, the research team discovered the molecular secret behind its power. When it attacks bacteria, cilagicin combines two molecules called c55-p and c55-pp, which bacteria use to build cell walls. It is well known that existing antibiotics can bind one molecule or another, but drug-resistant bacteria can still use the second molecule to maintain the cell wall. By solving these two problems at the same time, cilagicin may prevent the emergence of such resistance.
Although the study looks promising, the team cautions that much remains to be done before it can be used in humans. The researchers plan to optimize the compound and conduct more tests on animals against a range of other bacteria. But the most exciting thing is that cilagicin may only be the first of many new antibiotics discovered by this method.
Brady said: "this work is the best example of what can be found hidden in a gene cluster. We believe that we can now use this strategy to unlock a large number of new natural compounds. We hope this will provide an exciting new drug candidate library."
The study was published in Science 》In the magazine.